Our work on the discovery of novel anticancer peptides using AI approaches is published in the Journal of Chemical Information and Modeling. This is a collaborative work with the team of Henry Kwok, Chris Wong, and Lawrence Si from University of Macau. In this research, we developed an AI workflow to predict and rank translated sequences from bacterial genome for their anticancer activity. As a proof-of-concept, we applied the workflow to screen the genome sequence of Candida albicans and identified two novel anticancer peptides with potency of 3.75 and 56.6 uM. Our AI methods can be accessed from our webpage (AcPEP at CBBio Tools) for immediate prediction needs. However, for those wanting to massive screen some genomes, the codes are available for local installation. Cheers!
For more about this research, see our Research page or our full paper.

In silico ACP screening workflow for the identification of potential anticancer peptides for colorectal cancer from the genome sequence of C. albicans.
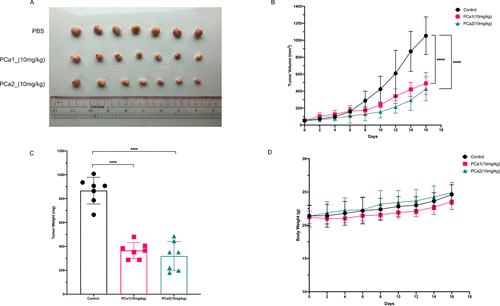
PCa1 and PCa2 inhibit the growth of colon cancer HCT116 tumors in vivo. (A) 5 × 105 HCT116 cells were subcutaneously injected into the nude mice to form tumor xenografts. Animals were treated with equal volumes of PBS, PCa1 (10 mg/kg), or PCa2 (10 mg/kg) by peritumoral injection every other day for 16 days as indicated. Images of HCT116 tumors were obtained at the end of the animal experiment. (B) Tumor volume (mm3) was measured every other day for 16 days. (C) Tumor weights were obtained at the end of the experiment. (D) Body weight of each mouse was recorded every other day for 16 days.
Leave a comment