To date, drug discovery has been a major challenge due to limited knowledge of disease and difficulty in identifying the right molecular targets and drug candidates. Our research focuses on developing computational methods to support drug discovery by innovating in the fields of bioinformatics, machine learning, deep learning, and optimization algorithms. We explore the therapeutic applications of peptides, including antimicrobial, anticancer, GPCR-interacting, and ion channel-interacting peptides. Our work involves modeling the molecular properties of these peptides and correlating them with the biological activities observed in experiments. Additionally, we focus on modeling and simulation of proteins to study their structures, dynamics, and functions. We have been involved in the development of membrane lipid force fields and more recently in the modeling of self-assembling monolayers on biochips. To facilitate virtual screening of drug candidates, we improve molecular docking programs by designing efficient search algorithms. This allows us to better predict the binding interactions between potential drug molecules and their target proteins, accelerating the drug discovery process.
Our multidisciplinary approach, combining expertise in computational biology, bioinformatics, and machine learning, enables us to tackle the complex challenges in drug discovery and development. Our ultimate goal is to contribute to the identification of novel therapeutic agents and the understanding of disease mechanisms.
Report of our recent research (MPU OpenDay 2026)

Discover Anticancer Peptides by Screening of Microbe Genomes using Artificial Intelligence
Web server: https://app.cbbio.online/acpep/home
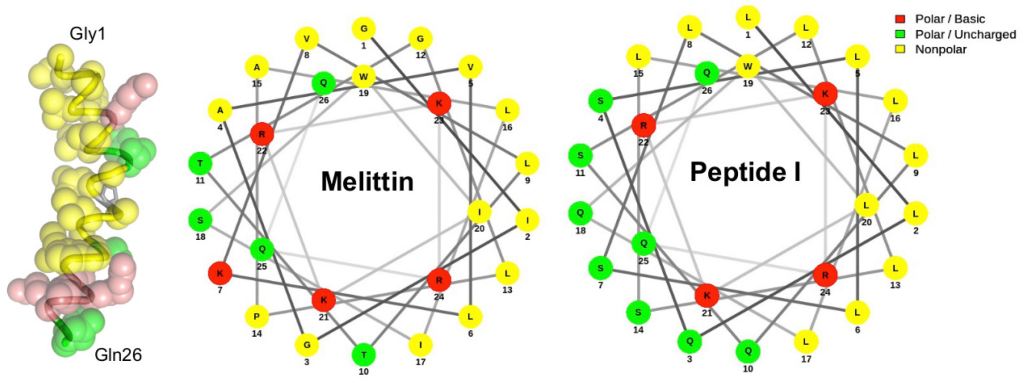
Cheong HH, Zuo W, Chen J, Un C-W, Si Y-W, Wong KH, Kwok HF, Siu SWI. Identification of anticancer peptides from the genome of Candida Albicans: In-silico screening, in-vitro and in-vivo validations. (accepted)
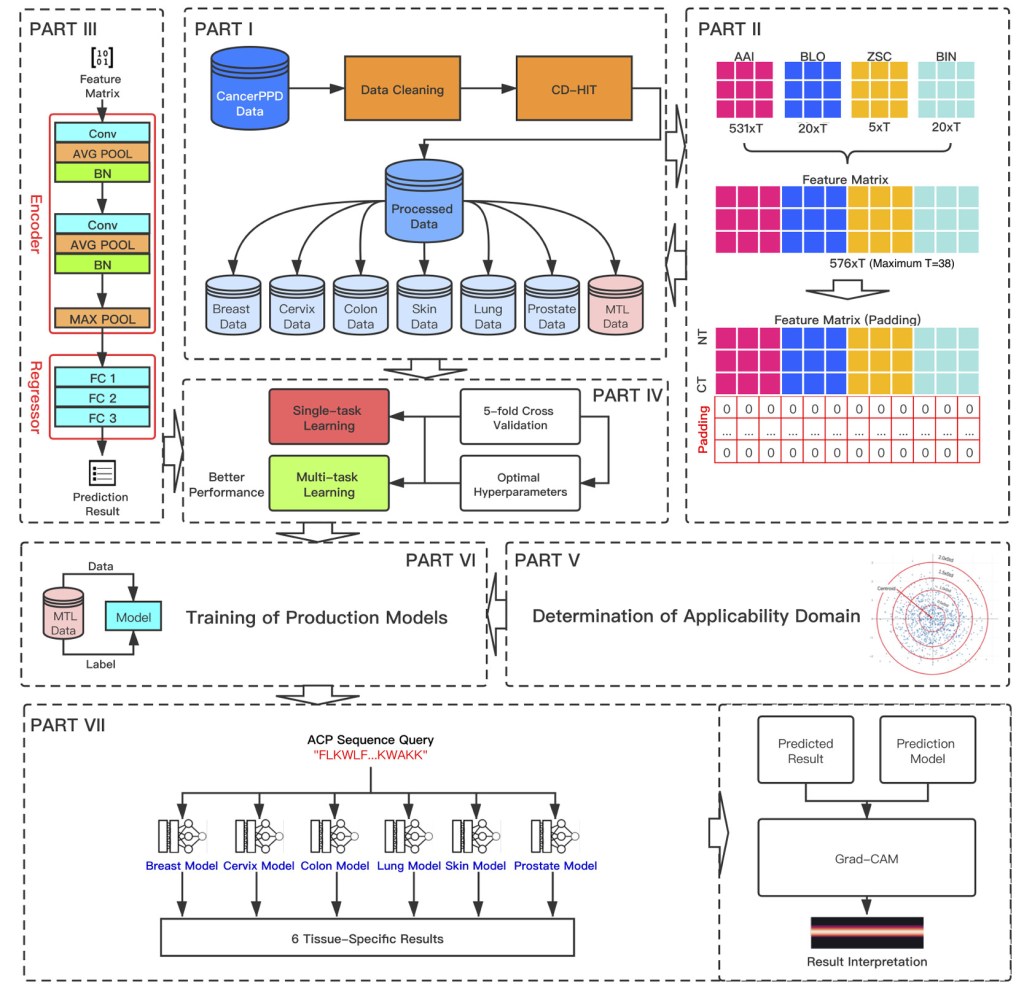
Chen J, Cheong HH, Siu SWI. xDeep-AcPEP: Deep learning method for anticancer peptide activity prediction based on convolutional neural network and multitask learning. J. Chem. Inf. Model. 2021, 61, 8, 3789–3803.
Anticancer peptides (ACP) open a new and promising avenue for the development of anticancer drugs with higher selectivity and fewer side effects than currently available therapeutics. By harnessing the power of machine learning techniques, we can identify novel ACPs in an accurate and efficient way using a data-driven approach. To facilitate the discovery of novel ACPs from natural sources, we use an in-silico screening workflow which allows us to extract potential ACP sequences from the genomes of organisms known to produce a diverse array of bioactive peptides.
Our workflow is a multi-step procedure which selects sequences with high anticancer potency and low toxicity. The workflow consists of a ACP classifier and a quantitative activity regressor (xDeep-AcPEP) for six types of tumor cells, including breast, colon, cervix, lung, skin, and prostate. Selected sequences are further filtered by toxicity predictors to generate a list of potential sequences. As a proof-of-concept, the workflow is used to identify novel ACPs for colorectal cancer from the genome sequence of C. albicans. Four candidate ACPs are tested in-vitro and in-vivo, demonstrate anticancer potent activity against colorectal cancer models while exhibiting low toxicity towards normal cells.
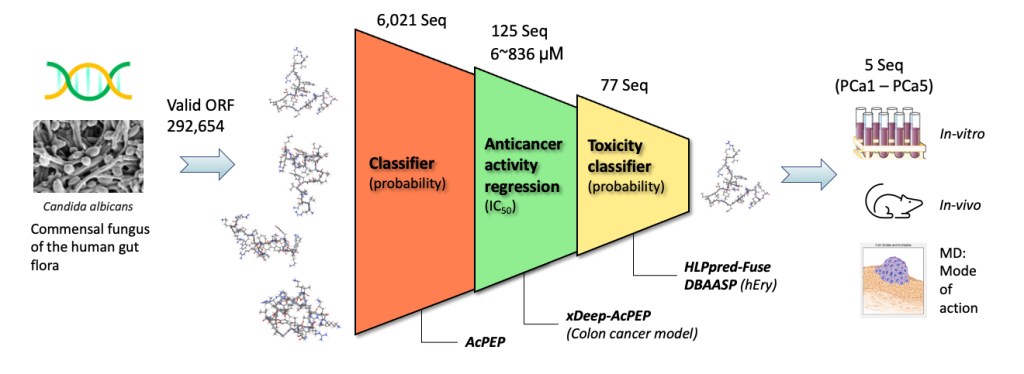
| No. | Sequence | Length/Charge | IC50 in HCT116 (uM) | IC50 in CCD-18-CO (uM) | Selectivity Index (SI) |
| PCa1 | RSLHCMNHLRLWIKLIWRILVKD | 23 / 4 | 3.75 | 15.28 | 4.07 |
| PCa2 | GKQAYQCLQMGVVMILKKLKK | 21 / 5 | 56.06 | 276.10 | 4.93 |
| PCa3 | ITNHNKKNIVLLKLHLILKL | 20 / 4 | 85.44 | 235.80 | 2.76 |
| PCa5 | YVDWFKCYFFPVILFNFCCRDI | 22 / 0 | 69.49 | 181.60 | 2.61 |
xDeep-AcPEP – Multitask Learning Model for Anticancer Activity Prediction
We develop a deep learning method based on convolutional neural networks to predict biological activity (EC50, LC50, IC50, and LD50) against six tumor cells, including breast, colon, cervix, lung, skin, and prostate. We show that models derived with multitask learning achieve better performance than conventional single-task models.
| Cancer Tissue | Model | MSE | PCC | p-value |
| Breast | MTL | 0.2059 | 0.7454 | 0.0019 |
| MTL with AD | 0.1832 | 0.8073 | ||
| Cervix | MTL | 0.2275 | 0.7677 | 0.0011 |
| MTL with AD | 0.1875 | 0.8322 | ||
| Skin | MTL | 0.2209 | 0.7014 | 2.62×10-06 |
| MTL with AD | 0.1661 | 0.7289 | ||
| Prostate | MTL | 0.2194 | 0.7762 | 0.0127 |
| MTL with AD | 0.2038 | 0.8179 | ||
| Lung | MTL | 0.2088 | 0.7947 | 0.0022 |
| MTL with AD | 0.1802 | 0.8370 | ||
| Colon | MTL | 0.1749 | 0.7189 | 0.0004 |
| MTL with AD | 0.1341 | 0.8285 | ||
| Average | MTL | 0.2096 | 0.7507 | |
| MTL with AD | 0.1758 | 0.8086 |
2) MTL with AD (MTL with applicability domain defined): This model provides more confident prediction, which is the model we deploy in the web server.
Discover Antimicrobial Peptides as Antibacterial Agents
Web server: https://app.cbbio.online/ampep/home
Publication: https://doi.org/10.1016/j.omtn.2020.05.006
Antimicrobial peptides (AMPs)[1] are a group of natural peptides that show promise as next-generation antibiotics due to their low toxicity to the host, broad spectrum of biological activity, including antibacterial, antifungal, antiviral, and anti-parasitic activities, and great therapeutic potential, such as anticancer, anti-inflammatory, etc. Most importantly, AMPs kill bacteria by damaging cell membranes using multiple mechanisms of action rather than targeting a single molecule or pathway, making it difficult for bacterial drug resistance to develop.
Remarkably, this DL method [2] is much more efficient than conventional ML methods [3] allowing massive screening of novel peptides.
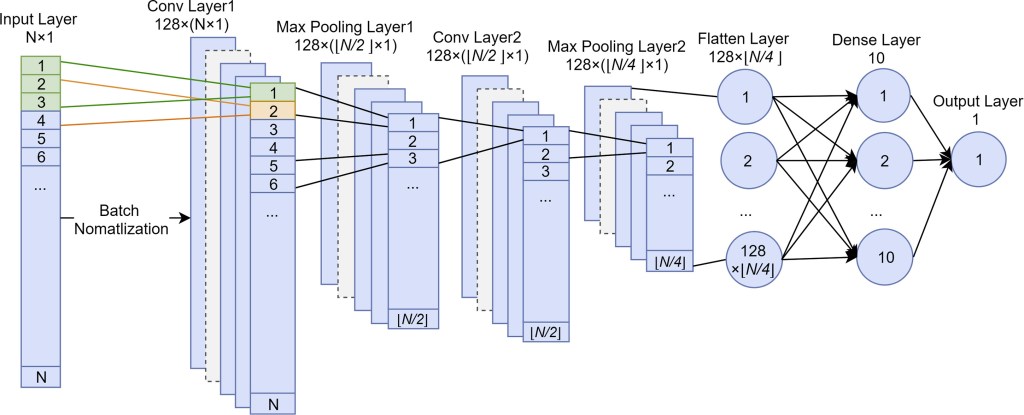
We developed a short-length (≤30 aa) AMP prediction method, Deep-AmPEP30, based on an optimal feature set of PseKRAAC reduced amino acids composition and convolutional neural network. The method was used to screen the genome sequence of Candida glabrata—a gut commensal fungus expected to interact with and/or inhibit other microbes in the gut—for potential AMPs and identified a peptide of 20 aa (P3, FWELWKFLKSLWSIFPRRRP) with strong anti-bacteria activity against Bacillus subtilis and Vibrio parahaemolyticus. The potency of the peptide is remarkably comparable to that of ampicillin.
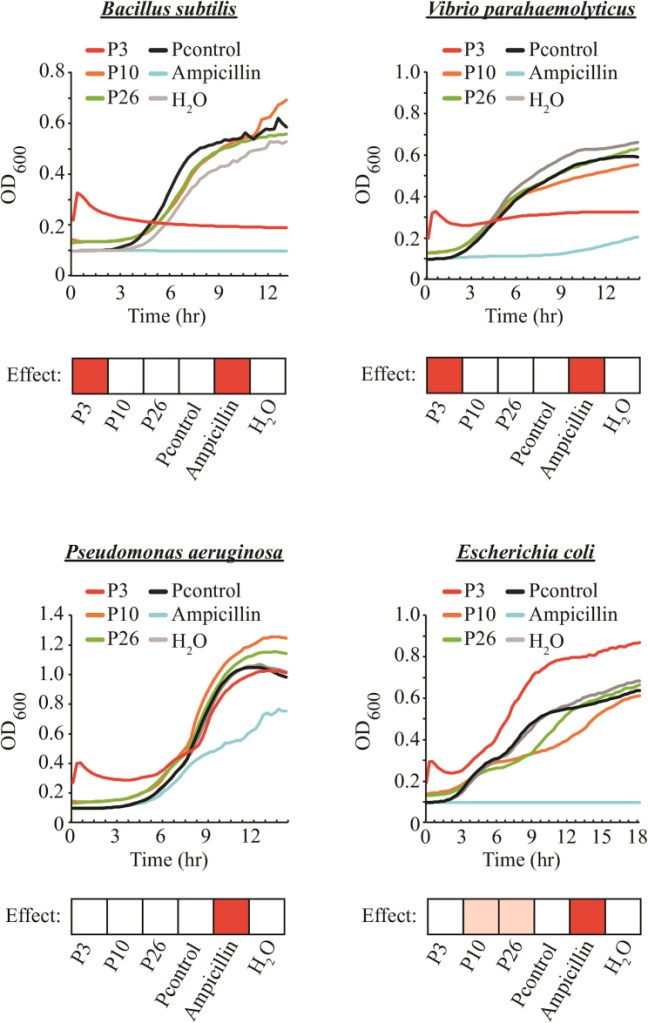
References
- Yan J, Cai J, Zhang B, Wang Y, Wong DF, Siu SWI. Recent Progress in the Discovery and Design of Antimicrobial Peptides Using Traditional Machine Learning and Deep Learning. Antibiotics (Basel). 2022;11(10):1451. Published 2022 Oct 21. doi:10.3390/antibiotics11101451
- Yan J, Bhadra P, Li A, Sethiya P, Qin L, Tai HK, Wong KH, Siu SWI. Deep-AmPEP30: Improve Short Antimicrobial Peptides Prediction with Deep Learning. Mol Ther Nucleic Acids. 2020 Jun 5;20:882-894. doi: 10.1016/j.omtn.2020.05.006.
- Bhadra P, Yan J, Li J, Fong S, Siu SWI. AmPEP: Sequence-based prediction of antimicrobial peptides using distribution patterns of amino acid properties and random forest. Sci Rep. 2018;8(1):1697. Published 2018 Jan 26. doi:10.1038/s41598-018-19752-w